Step 1
Select the organism of interest (Human or Mouse).
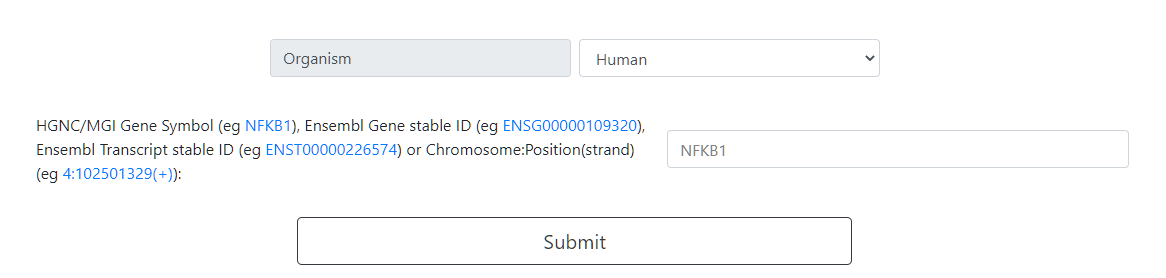
Type the gene of interest. The Ensembl codes of a gene and its transcripts can be used as input as well.
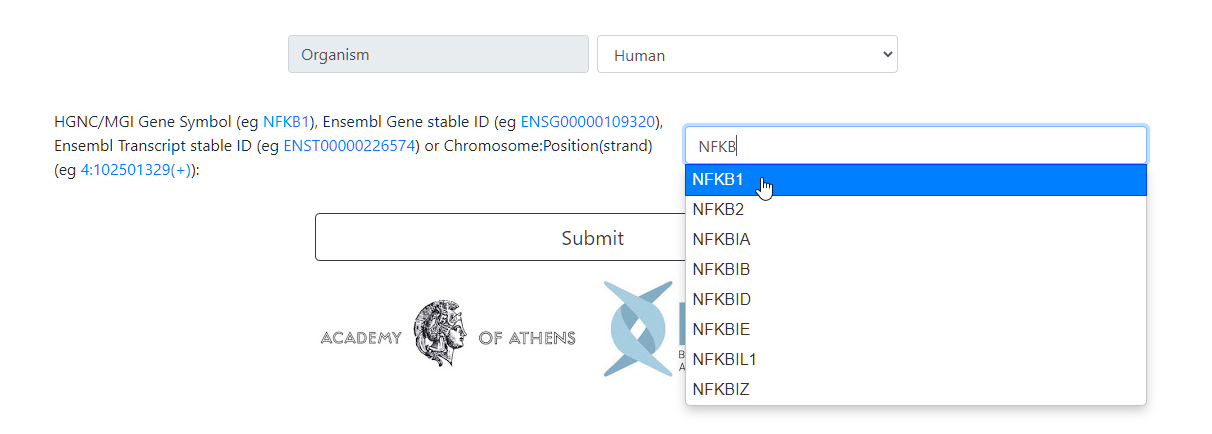
A custom specific region can be typed. The format has to be precise Chr:Location(Strand). This tool uses the genomic location numbers of the Ensembl 90 release, Homo sapiens hg38 and Mus musculus mm10 .
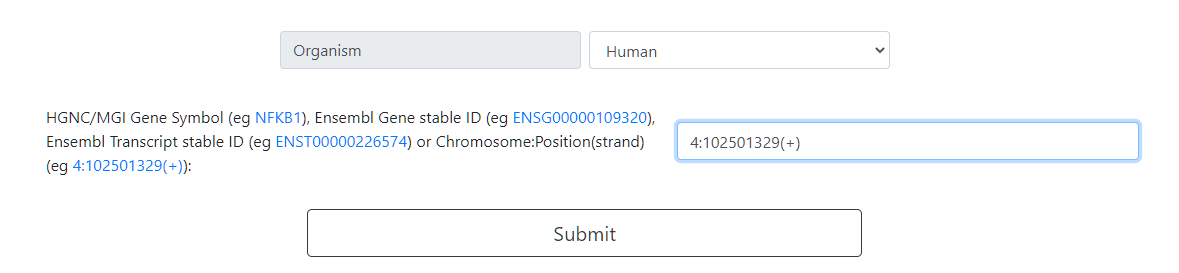
Click on Submit to Continue.
Step 2
Click on a TSS to Continue. The rest of the links redirect to the corresponding entries in GeneCards or Ensembl.
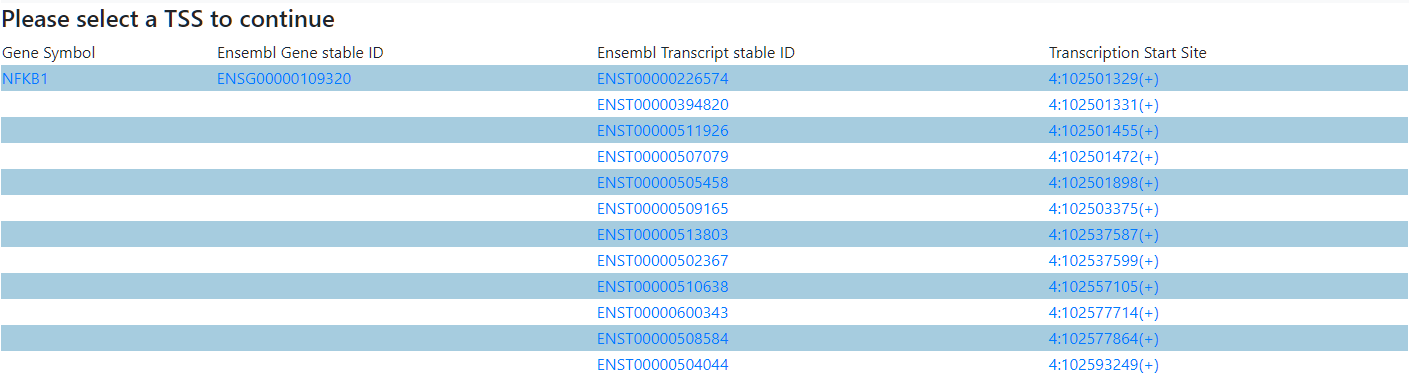
Step 3
Select the categories of the cell types that will be examined. At least one option of each category must be selected.
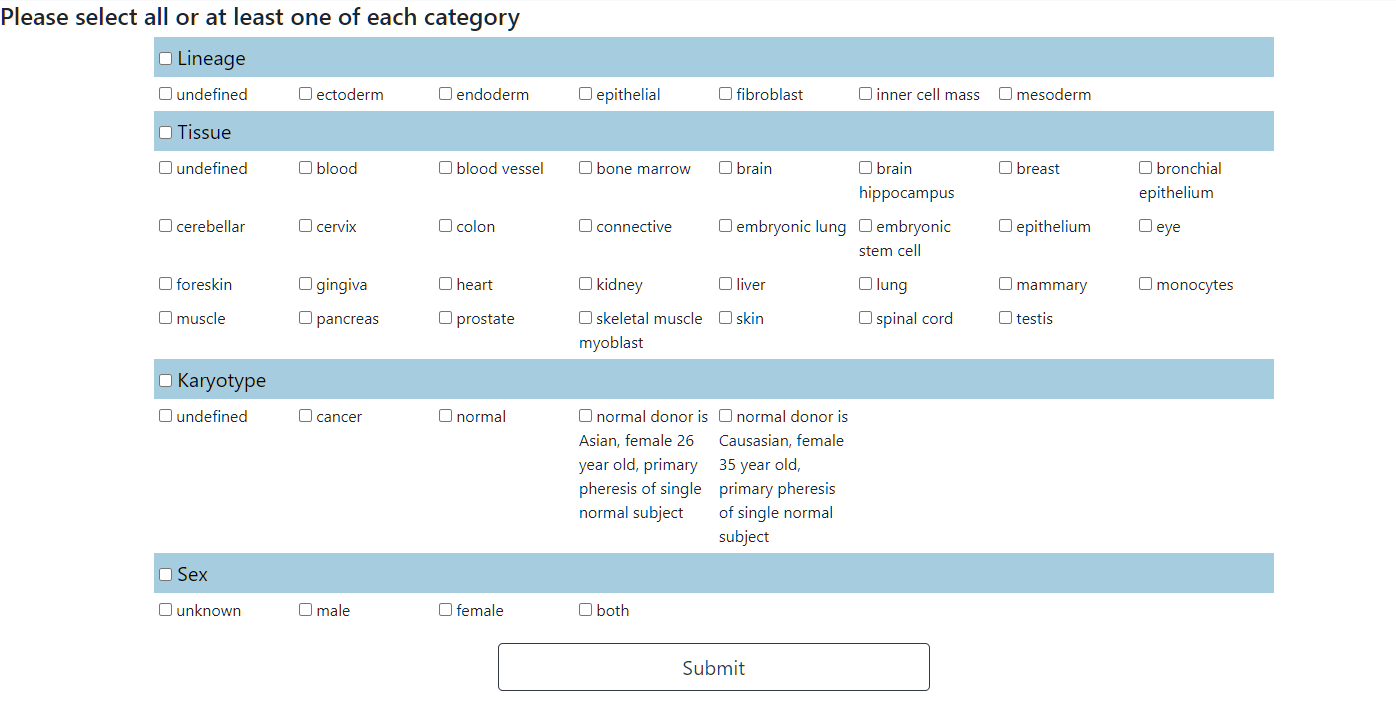
Step 4
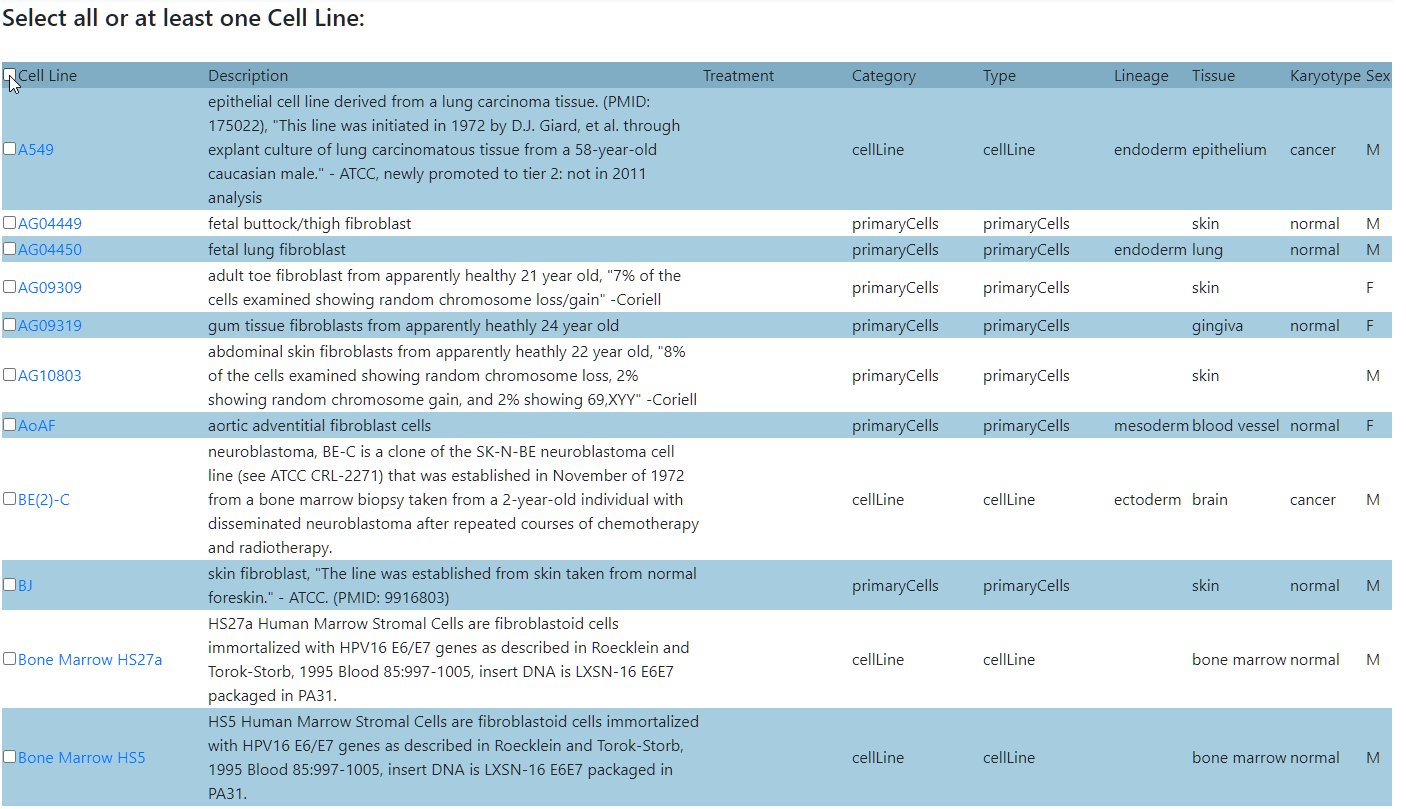
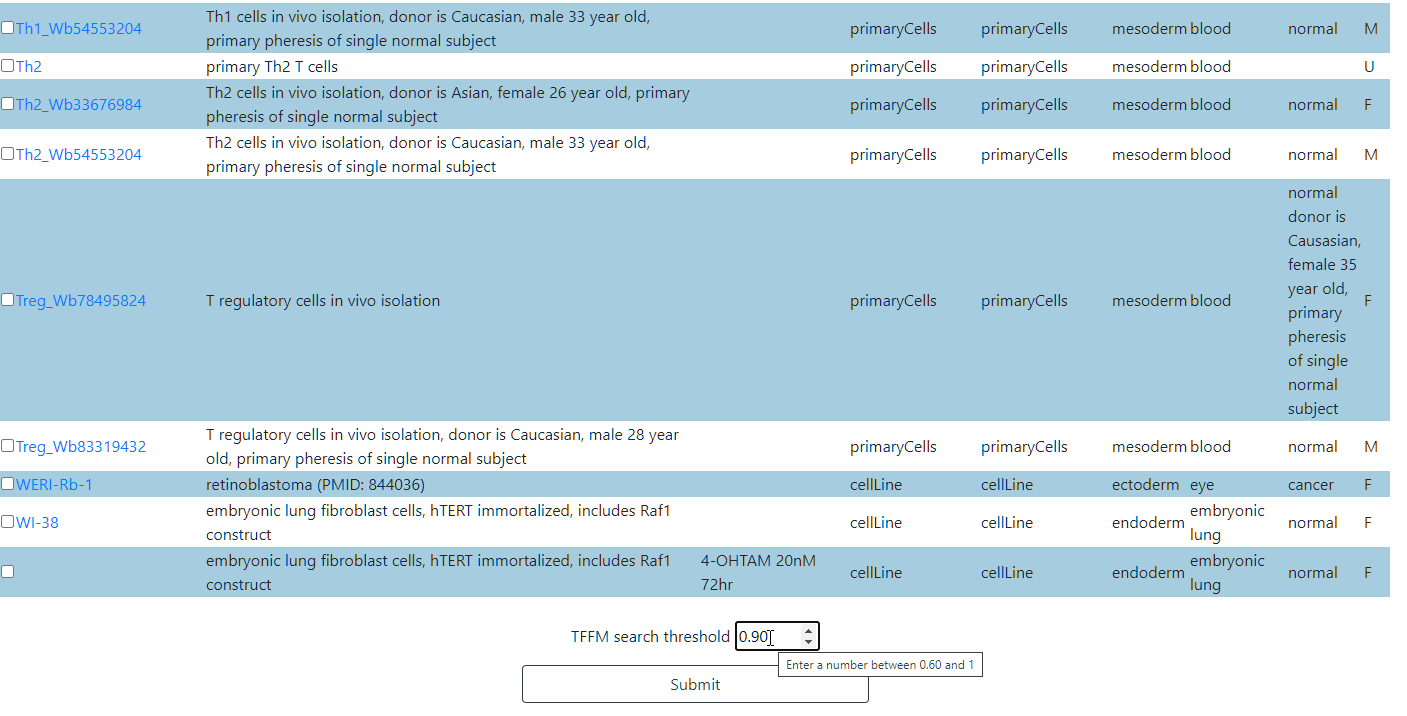
Select one or more cell types from the selected categories.
There is also a TFFM threshold field where the threshold of the search algorithm can be adjusted. The default value is 0.90.
Click on Submit to Continue.
Step 5
On the results page, there is the pairwise alignment of the region where promoter analysis is made. The TSS is coloured red and above it, an arrow shows the orientation of the transcription. The predicted TFBS are also displayed above each alignment block.
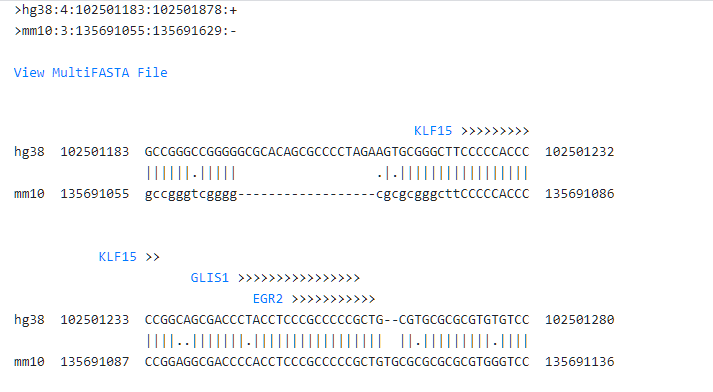

Below the pairwise alignment details of the TFBS predictions can be displayed.
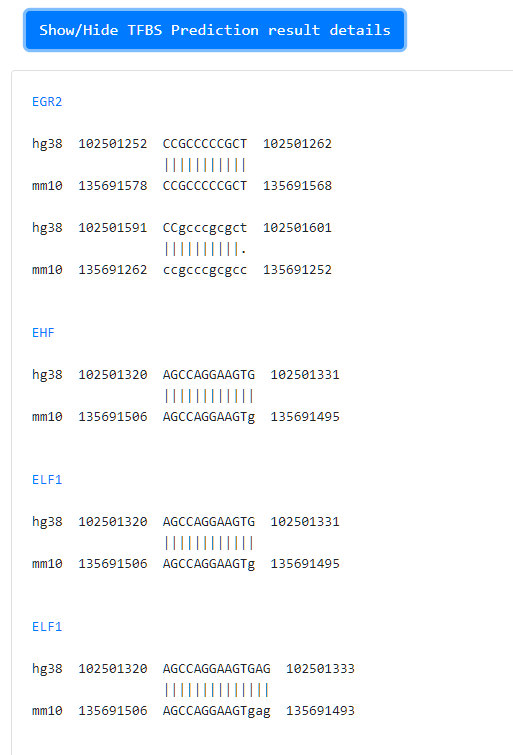
Any of the TFs can be clicked to be redirected to its JASPAR entry page.